ABSTRACT
In the academic process, comprehension and analysis of literature is essential, however, time-consuming. Reviewers may encounter difficulties in identifying relevant literature, given the considerable volume of available texts. It is arduous not only for starting PhD students, but also for any researcher learning about a new field (called here "domain learners"). To address this issue, we present an automated framework to assist in the literature review process. Through the application of topic modelling of academic articles, our framework encourages senior researchers within a specific field to act as experts to contribute to the labelling of topics. Further to this, domain learners can benefit from visualisation tools intended to assist in the comprehension of vast amounts of academic texts. Our approach allows reviewers to identify the topics, trends as well as relations between topics in a given research field. We also accompany this method with a tool that we provide open source. For illustration, we apply here our method to a case-study of biological texts, specifically texts related to human protein kinases. To further enhance the educational capabilities of our approach, we perform triangulation of external biomedical databases, to illustrate how our multi-pronged approach can provide a comprehensive understanding of the research domain.
Keywords
1. INTRODUCTION
The process of understanding academic literature is a time-consuming process for both students and professionals. Thus, there is a necessity to enable the process of quickly comprehending such literature. Furthermore, limitations to the amount of work which can be analysed by an individual or team may be encountered, due to the time it takes to understand each item of literature. This is of particular relevancy to the biomedical field, where estimates in 2016 proposed that the field observes 3 new publications per minute uploaded to the PubMed database alone [1]. Moreover, the identification and recognition of evidence and named entities within full-text biological literature - the Curator Assistance Problem [1], is limited by the requirement for manual analysis of text by experts within the field, using their own knowledge and intuition.
To address these issues, we propose an automated approach to assist in the literature review process in biomedical literature. By leveraging recent advancements in topic models, we seek to provide novel benefit to the academic and research process when learning about a certain new research area. Further to this, to address the Curator Assistance Problem [1], we perform rule-based identification of human-related protein kinases within the literature, for automatic triangulation of multiple external data sources, to assist in comprehension of research. We provide access to the case-study for reproducibility 1. Our research aims to address the following research questions: 1. How may topic modelling algorithms be applied to assist in the comprehension of large volumes of academic data? 2. How may semantic similarity measures be leveraged to identify inter-topic relationships of identified literature? 3. Can rule-based extraction of named biomedical resources contribute to the comprehension of automatically generated literature topics? 4. Can scientific language-oriented embedding models provide improvements to the perceived accuracy of topic-modelling, when analysed by biological domain-experts?
The main contributions of this paper are thus: (1) Provision of a novel framework for the analysis of literature topics by accounting for semantic relationships and temporal trends of identified topics. (2) A capability for the crowdsourcing [2] of domain experts for the labelling and filtering of topics within the literature. Experts such as educational tutors or senior researchers may contribute to the labeling of identified topics based upon their own knowledge. Subsequently, domain learners may then benefit from the visualisation tools provided to assist in their learning on the subject. (3) Triangulation of multiple external data sources for the extraction and linking of named kinases present within literature, to assist in the expert labelling and comprehension of topics within the literature. Although this is a case-study based upon literature related to protein kinases, in a collaboration with bioscientists, this robust approach allows for further generalisation, and could be applied to the analysis of literature from any domain. (4) An unsupervised evaluation of the performance of scientific-domain transformer embeddings, compared to document embedding approaches when applied to document clustering for the identification of topics within scientific literature.
2. RELATED WORKS
As a data-intensive field, there has been an emergence of numerous biological data-sources of various quality and provenance. However, due to a lack of a standard ontology with fine granularity, it is difficult to conduct data mining on those heterogeneous biology data. Despite efforts to normalise data formats and collect observations in databases, a large amount of information relevant to biology research is still recorded as free text in journal articles and in comment fields of databases [3]. Therefore, retrieving information from papers and merging it with existing biological databases can provide a crucial foundation for data-mining in bioinformatics [1]. To address this, works have been proposed for the extraction of entities within biological texts, including RegulonDb [4], which applied rule-based identification of regulatory interactions in Escherichia coli, with the results demonstrating a rule-based approach sufficient in identifying 45% of all named entities when compared with a manual extraction approach. Extraction of genetic and protein interactions was performed by BioC-BioGRID, which released a corpus of 120 full-text articles with biological and molecular entity annotations [5]. Based upon these problems and subsequent approaches, [1] proposed the Curator Assistance Problem. This problem proposes four objectives, which were defined given a full-text biological research paper: 1. The recognition of evidence described in an article, compared to information in other articles. 2. The recognition of evidence which is supported by experimental data, compared to hypothetical of vague statements. 3. The distinction between statements on layout, compared to statements of results. 4. The recognition of negative statements.
No research was identified relative to the automated literature review process for biomedical data. However, in the wider domain, one framework has been demonstrated to identify relevant papers for a literature review based upon an input of seed papers [6]. This approach applied supervised learning classifiers, which were trained upon reference lists of existing papers. From an unsupervised perspective, [7] applied topic modelling to the task of conducting an exploratory literature review through the use of Latent Dirichlet Allocation (LDA). The approach required application of the elbow curve methodology within an exhaustive search, to determine the optimum number of topics within the literature. In contrast, a recent topic modelling approach, Top2Vec [8], has presented advantages over the application of LDA, through the elimination of the need to manually or exhaustively define the optimal topic number. Applications of Top2Vec to assist in literature analysis were presented in [9], through a case-study review of research in Intelligent Tutoring Systems. However, the approach required the manual analysis of resulting topics prior to the generation of visualisation and topic labels. Our study is based on this, but is expanding it in several ways. Importantly, we propose a framework for the e-learning domain, and illustrate it through an interface, to permit the crowdsourcing [2] of topic labelling on a large scale, as well as provide an expanded suite of tools to assist with learning. By creation of this framework, we are able to demonstrate the generalisable characteristics of the approach, such that it may be applied to any domain of literature, and may be subsequently enhanced through the implementation of domain-relevant features, such as the protein databases applied for our case-study. The resulting implementation of domain-relevant data triangulation may enhance the information available to researchers by providing supplementary information to aid in filtering and identification of relevant literature to a domain-user.
3. METHODOLOGY
The proposed framework (as summarised in Figure 1) entails three components. Firstly, topic modelling is performed on the academic literature that is identified as relevant to the case-study domain through the use of the API resource. Following this, senior researchers are presented with a interactive topic labelling and visualisation analysis tool, which allows them to determine whether a topic is relevant to their literature search, as well as assign a title for that topic. Finally, this visualisation analysis tool can facilitate the comprehension of relevant literature of the domain learners. To illustrate the framework, here, based on the full-text of documents assigned to a topic, extraction of human-protein-kinase terms (the topic of the literature review) is performed by rule-based extraction of kinase names. Also, in terms of the actual implementation, an internal relational database provides external links to the external resources – here, those coming from UniProt Knowledgebase (UniProtKB) [10] and InterPro [11], well-known protein databases. The resulting interface may be applied as a framework for learning through the crowdsourcing of domain experts within a field for the labelling and filtering of literature topics. Expertly annotated topics may then be provided to learners or experts, who may apply the visualisation tools provided to assist in their learning and comprehension of the literature.
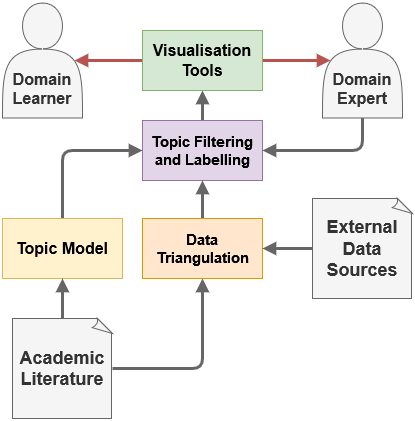
3.1 Data Sources
A full-text representation of publications is provided through the PubMed API, which ensures to capture the full semantic context of a text if it is analysed by a compatible language model for variable length document-embeddings. Two additional sources were selected for the triangulation of kinases identified within the literature, with these being the UniProtKB [10] and Interpro [11] databases. UniProtKB is a database of protein sequences and functional information, which includes information relevant to specific proteins, their function, organism, presence in biological processes, and relevant literature. InterPro serves as a similar platform, however includes hierarchical family classifications of protein entries as well as domains and functional sites found in proteins.
For the case-study, a list of 624 human protein kinases was firstly obtained from a kinase database [12, 13]. A comprehensive list of human proteins with links to their respective InterPro IDs was obtained from UniProtKB (Swiss-Prot, Jan 2014 version) [10]. Matching of protein kinase names to gene names including synonyms for proteins from UniProtKB was performed. The resulting 357 protein kinases that successfully matched to UniProtKB entries were used in this study. This entailed the names of relevant genes, kinase names and kinase families. Further to specific kinase names (e.g. AATK1), the data included full-named entities relevant to a kinase. Relevant to the given example, this would be Apoptosis-associated tyrosine kinase 1. This list of shortened and full terms was applied in an automated literature search via the PubMed API [14]. A total of 19,258 records were returned in this manner. Following this, filtering was performed, to ensure the elimination of any noise introduced into the dataset during the search stage. This was performed via filtering out publications that failed to contain Medical Subject Heading (MeSH) terms related to Humans. Following elimination of non-human-related subjects, 14,631 documents remained.
3.2 Topic Modelling
For the identification of topics within our literature, the Top2Vec [8] algorithm, which leverages the HDBSCAN [15] algorithm, is applied to identify dense clusters of document embeddings, and determine these to be topics. The algorithm automatically generates human-readable labels, based upon the top-scoring topic-words for each topic. In addition, a domain-expert may manually assign a title for each topic on the topic labelling screen. The use of HDBSCAN presents a unique opportunity when clustering, given that HDBSCAN may label documents determined to be noise [15]. This provides the benefit of eliminating documents which do not directly fall within a topic.
3.3 Literature Analysis Toolkit
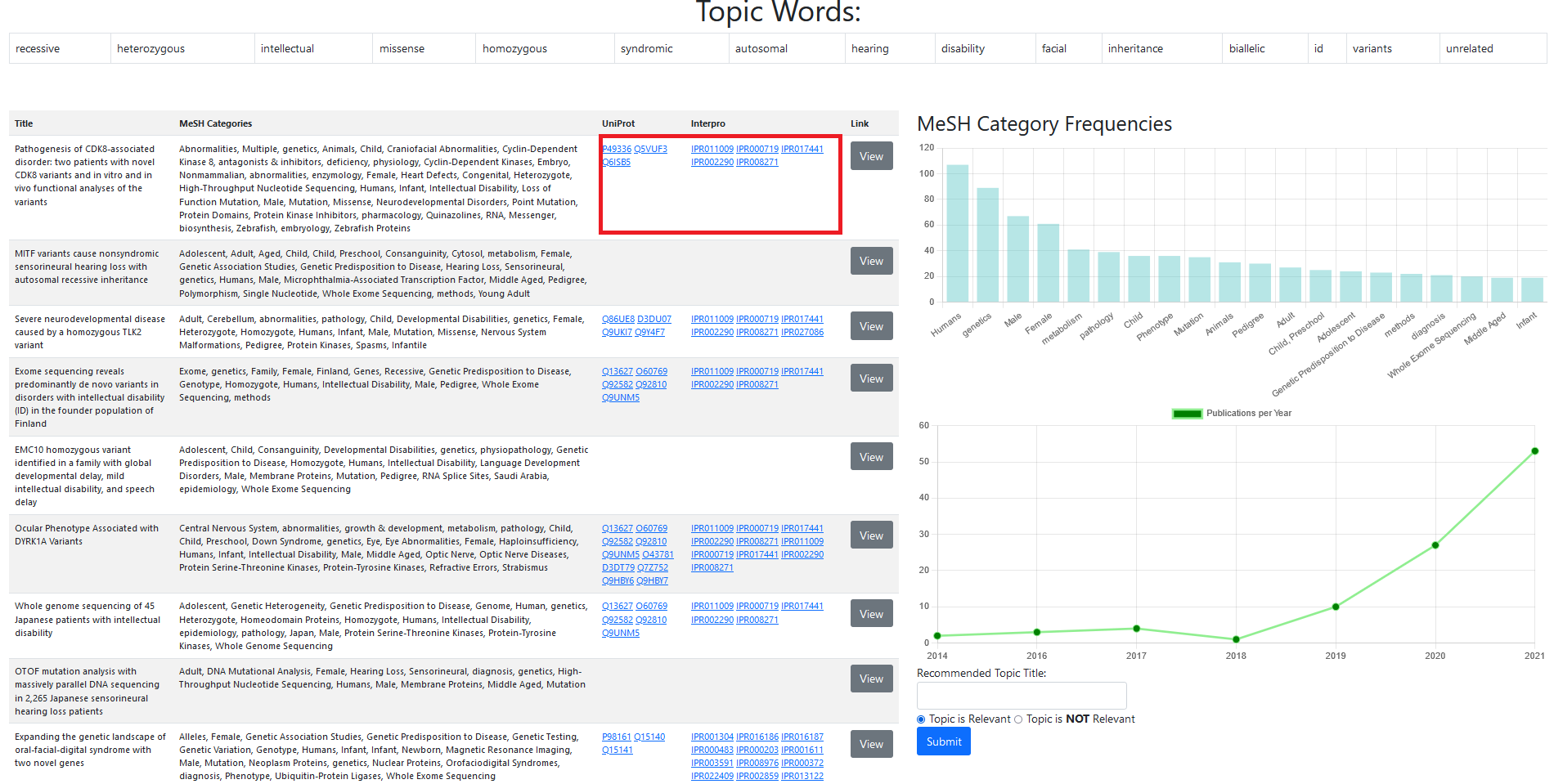
The main contribution of our approach centers around the development of a simple user interface for evaluating the results of literature topic analysis, and subsequent deeper analysis. Researchers may find difficulty in the application of algorithmic analysis approaches, as these require a degree of knowledge of programming and an understanding of the underlying algorithms applied. Therefore, through providing an interactive user interface, we ensure that researchers from the biomedical domain may analyse literature without the need to spend time learning the underlying system. Results from the topic modelling of the literature are presented to the domain-expert in a simple landing page, which provides a table format of top-scoring words for each topic, the manually assigned label, and relevancy classification which can be manually defined. By clicking on the Edit Label button, domain-experts may view a detail page (as shown in Figure 2), featuring top-scoring publications assigned to a topic, the top-scoring words for the topic, and a bar graph visualisation, detailing the total frequency of MeSH terms for publications assigned to that topic. Providing these features presents domain-expert with an opportunity to decide the title of the topic through a number of means. For example, MeSH subject headings can provide the frequency in which certain diseases are mentioned within documents in a topic. Further to this, by extracting mentions of human protein kinases within full-text articles, we facilitate the triangulation of the InterPro [11] and UniProtKB [10] databases, any may provide domain-expert with external links (as shown in the highlighted box in Figure 2) to related resources relative to a given publication. Domain-expert may also view the full publication at the place of publishing through a hyperlink, so that individual publications they find useful through the above analysis methods may then be read further.
Further academic features are provided to users (both domain experts and learners) through an analysis of the semantic relationships within topics, following the same methodology presented by [9]. Semantic relationships are generated through the calculation of the cosine similarity between all combinations of topic vectors, where a topic vector is defined as the average of all document embeddings belonging to that topic. We define each topic as a node, with the top three highest scoring topic similarity pairs (edges) for each topic being presented in the graph visualisation. Nodes are presented with at least three edge relationships. However, some nodes may have more than three edges. For instance,as shown in Figure 3, the node "DNA repair" displays the top 3 scoring edge relationships with "Genetic disorders," "Cancer genetics" and "Phosphorylation profiling". However, For the "Radiation effects" and "Alternative splicing" nodes, the "DNA repair" edge relationship is within the top-3 ranking for the nodes respectively, hence "DNA repair" demonstrating 5 edge relationships.
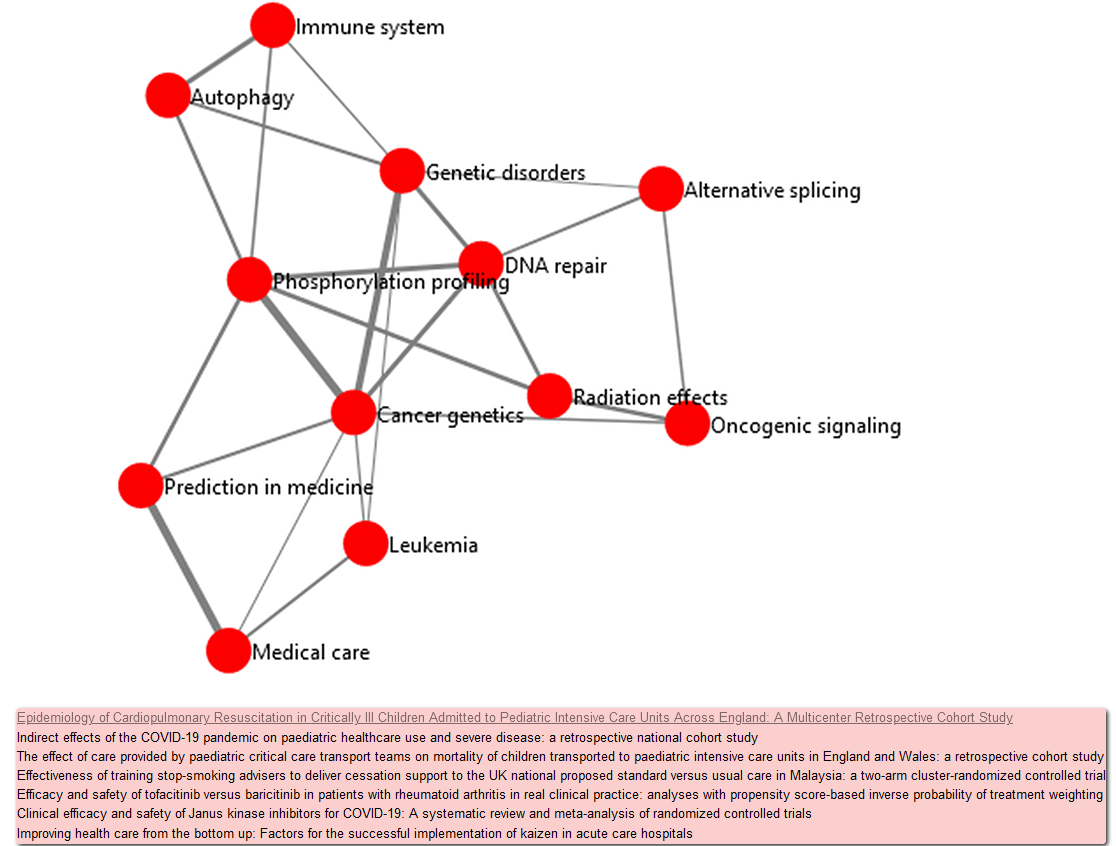
The topic-relationship graph is computed when initialising the system, and store this in a serialised format. Edge thickness is a reflection of the cosine-similarity score between two nodes, for easy understanding of the strength of the relationship between those topics, with a tooltip presenting the similarity score when the user hovers their mouse over an edge. From this view, users may also further expand a topic, navigating to a view of the top-scoring academic texts that have been assigned to that topic based upon the distance of the publication to the topic centroid, by right-clicking on a node. For example, the pink panel on the bottom of Figure 3 is displayed when right-clicking the node "Medical care". This panel provides links of the paper directly to the external resources. Users may choose to view all identified topic-relationships from the topic modelling process, or only those that have been manually labelled as relevant.
4. CASE STUDY IN BIOLOGY - HUMAN PROTEIN KINASES
As a case study, the toolkit generated 279 topics from texts of publications related to the 357 human protein kinases. A representative list of associations, for each topic, is provided and contains the top twenty highest scoring topic-words, top ten individual publications, and MeSH category frequencies of publications assigned to the topic. For example, the topic (shown in Figure 2) is provided with the following information: relevant topic words such as "recessive," "heterozygous," "inheritance," "missense," "homozygous," "syndromic," "autosomal," "disability," and "variants"; titles of individual publications with phrases such as "neurodevelopmental disease caused by a homozygous TLK2 variant," "variants in disorders with intellectual disability," and "phenotype associated with DYRK1A variants"; and MeSH categories such as "Humans" and "Genetics" with the highest frequency in the assigned publications. Based on this information, the topic can be labeled as “Genetic disorders” associated with human protein kinases. Moreover, links to UniProtKB and InterPro databases are listed for specific kinases included in each individual publication. For example, the publication titled "Pathogenesis of CDK8-associated disorder: two patients with novel CDK8 variants and in vitro and in vivo functional analyses of the variants” is provided with the following links: the entry of the kinase CDK8 in UniProtKB; and the entries of domains, binding-sites, etc. found in CDK8 such as “Protein kinase domain," "Protein kinase, ATP binding site," and "Serine/Threonine protein kinases active-site" in InterPro. Such external databases provide detailed information such as biological functions, disease association, sequences, and domain architectures of protein kinases along with links to further explore other databases, should such a need arise for the user.
The toolkit also easily visualises relationships among topics based on their semantic similarity. To demonstrate its efficacy, the bio-experts in our team selected 12 topics that have at least six out of ten individual publications with links to protein kinases in UniProtKB, and then manually labeled the topics as shown in the previous paragraph. The resulting graph network (shown in Figure 3) generated from these topics has 12 nodes with the assigned labels and edges with different thickness that reflects the similarity between the linked topics . The node "Genetic disorders" is strongly connected to the nodes of "Cancer genetics" and "DNA repair." This is consistent with the idea that both genetic disorders and cancer genetics involve genetic changes and genomic technologies. Similarly, DNA repair defects are associated with certain genetic disorders. For example, ATM and ATR are DNA repair-associated serine/threonine kinases and their genetic mutations can cause hereditary disorders, ataxia telangiectasia and Seckel syndrome, respectively. Thus, it is reasonable to include both ATM and ATR in topic-words for the topic "DNA repair." Another strong connection visualised by the network is between "Medical care" and "Prediction in medicine," which aligns with the clinical aspect of treatment. The visualisation enables the users to easily explore major topics and their relationships. Collectively, the toolkit allows users to navigate a new field through vast amounts of literature in an efficient way.
5. CONCLUSIONS
In this work, an automated literature review framework is proposed for the analysis of large volumes of literature. The resulting topics from topic modelling of academic literature within specific domain, are provided via an interactive UI tool, which may allow the manual analysis and filtering of resulting topics, and exploration of entailing literature. Based on a case study on the biomedical domain of human protein kinases, we provide a further contribution to the comprehension of literature through the triangulation of biological relations present in identified publications. From an educational data mining perspective, our tool achieves the goal of grouping large amounts of academic text into understandable topics, and allows for crowdsourcing of expert-knowledge for the labelling of topics identified in the literature. The resulting framework achieves two objectives by serving as a base literature review assistance tool, or as an e-learning utility to allow expert analysis of topics, before providing the results to learners. By the inclusion of external databases UniProtKB and InterPro, we provide assistance in the extraction of named kinases within individual items of literature, allowing users to easily read further into the identified kinases present. Overall, this framework incorporates topic modelling with a crowd-sourcing approach, achieving the goal of expediting the task of analysing large volumes of literature.
6. REFERENCES
- Jacques Nicolas. Artificial Intelligence and Bioinformatics, pages 209–264. Springer International Publishing, Cham, 2020.
- Larissa Hammon and Hajo Hippner. Crowdsourcing. Business & Information systems engineering, 4(3):163–166, 2012.
- Lynette Hirschman, Jong C Park, Junichi Tsujii, Limsoon Wong, and Cathy H Wu. Accomplishments and challenges in literature data mining for biology. Bioinformatics, 18(12):1553–1561, 2002.
- Carlos Rodríguez-Penagos, Heladia Salgado, Irma Martínez-Flores, and Julio Collado-Vides. Automatic reconstruction of a bacterial regulatory network using natural language processing. BMC bioinformatics, 8(1):1–11, 2007.
- Rezarta Islamaj Doğan, Sun Kim, Andrew Chatr-aryamontri, Christie S. Chang, Rose Oughtred, Jennifer Rust, W. John Wilbur, Donald C. Comeau, Kara Dolinski, and Mike Tyers. The BioC-BioGRID corpus: full text articles annotated for curation of protein–protein and genetic interactions. Database, 2017, 01 2017. baw147.
- Jason Portenoy and Jevin D. West. Constructing and evaluating automated literature review systems. Scientometrics, 125(3):3233–3251, Dec 2020.
- Claus Boye Asmussen and Charles Møller. Smart literature review: a practical topic modelling approach to exploratory literature review. Journal of Big Data, 6(1):93, Oct 2019.
- Dimo Angelov. Top2vec: Distributed representations of topics. CoRR, abs/2008.09470, 2020.
- Ryan Hodgson, Alexandra Cristea, Lei Shi, and John Graham. Wide-scale automatic analysis of 20 years of its research. In Alexandra I. Cristea and Christos Troussas, editors, Intelligent Tutoring Systems, pages 8–21, Cham, 2021. Springer International Publishing.
- The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research, 49(D1):D480–D489, 11 2020.
- Matthias Blum, Hsin-Yu Chang, Sara Chuguransky, Tiago Grego, Swaathi Kandasaamy, Alex Mitchell, Gift Nuka, Typhaine Paysan-Lafosse, Matloob Qureshi, Shriya Raj, Lorna Richardson, Gustavo A Salazar, Lowri Williams, Peer Bork, Alan Bridge, Julian Gough, Daniel H Haft, Ivica Letunic, Aron Marchler-Bauer, Huaiyu Mi, Darren A Natale, Marco Necci, Christine A Orengo, Arun P Pandurangan, Catherine Rivoire, Christian J A Sigrist, Ian Sillitoe, Narmada Thanki, Paul D Thomas, Silvio C E Tosatto, Cathy H Wu, Alex Bateman, and Robert D Finn. The InterPro protein families and domains database: 20 years on. Nucleic Acids Research, 49(D1):D344–D354, 11 2020.
- Gerard Manning, David B. Whyte, Raquel Martinez, Tony Hunter, and Sucha Sudarsanam. The protein kinase complement of the human genome. Science, 298:1912 – 1934, 2002.
- Protein kinases, kinomes and evolution, at kinase.com.
- Entrez programming utilities help, Jan 1970.
- Ricardo J. G. B. Campello, Davoud Moulavi, and Joerg Sander. Density-based clustering based on hierarchical density estimates. In Jian Pei, Vincent S. Tseng, Longbing Cao, Hiroshi Motoda, and Guandong Xu, editors, Advances in Knowledge Discovery and Data Mining, pages 160–172, Berlin, Heidelberg, 2013. Springer Berlin Heidelberg.
1https://github.com/ryanon4/topic_labelling_tool
© 2022 Copyright is held by the author(s). This work is distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0) license.